Home > Research Achievements > GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis
GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis
(Nature, 459:455-459, 2009)
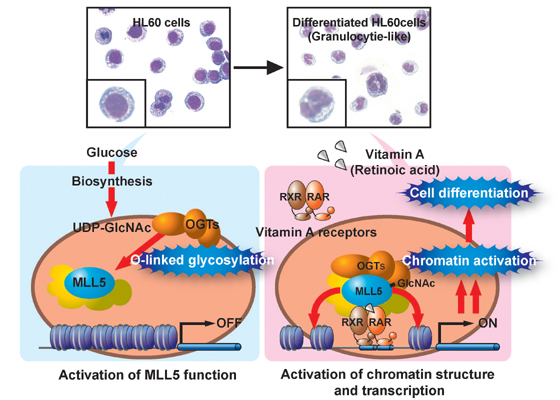
Differentiation results from a cell-specific gene expression patterning (aloso known as epigenome). This ‘pos-genomic’ era, a major question in biology is ‘What is the molecular mechanism of epigenome, to determine cell-specific chromatin structure and transcriptional outcome?’ A team led by Professor Shigeaki Kato of the Institute of Molecular and Cellular Biosciences, The University of Tokyo, they found that nuclear O-linked gylcosylation of a histone methyltransferase to facilitate retinoic-acid induced granulopoiesis of HL60 cells, traditional and well-known cellular differentiation model.
Fujiki R. et al. applied biochemical purification to this model for identifying an epigenetic regulator through activation of nuclear RA receptors (RARs). We purified a novel histone lysine methyltransferase (HKMT), MLL5, by using bacterially prepared RAR as bait, and found that MLL5 co-activated RAR-mediated transcription through its HKMT activity. O-linked N-acetyl-glucosamine (O-GlcNAc) transferase (OGT) is contained in HKMT-active MLL5 complex and activated MLL5 through GlcNAcylation of its catalytic SET domain. Further, as extracellular glucose level relevant to nuclear O-GlcNAc level, high-glucose condition dramatically effect on RA-response of HL60 cells. Post-translational glycosylation have been believed important in moleclular action of cell surface or extracellular proteins. In this aspect, we could propose the novel molecular mechanism linking glycobiology and epigenetics. The studies were published in May 21st 2009 issue of NATURE.Program member
Shigeaki Kato (Institute of Molecular and Cellular Biosciences, University of Tokyo)
Copyright (C) 2007-2009 All rights reserved tokyo-u.ac.jp
